Pharmaverse: Packages for clinical reporting workflows

Pharmaceutical companies spend significant money and resources trying to bring therapeutics to market faster. Regulatory submissions often entail similar and repetitive activities, and innovations in this area mean tremendous gains in efficiency and quality. Around 2009, the industry started to change as open source provided an easier, more collaborative, and more efficient way to tackle these tasks.
In August 2018, pharmaceutical companies gathered at Harvard University to discuss R and open-source tooling for drug development at R in Pharma. For years, industry leaders have been scaling their clinical trial workflows with tools like Shiny, Quarto/R Markdown, and the tidyverse. These core packages improve and automate data management and reporting, and they were the main focus of the R in Pharma conference from 2018-2020.
On top of these core tools, organizations create custom functions and packages that aid in their unique requirements. One of the most popular questions at 2018 R in Pharma was, “Is the package code available or on CRAN?”. While cross-enterprise, open-source collaboration is a common theme in pharma (in projects such as Bioconductor, R in Pharma, R Validation Hub, and Cross Pharma High Performance Computing), having availability to clinical workflow packages and code was often locked behind company firewalls.
This all changed at the 2021 R in Pharma conference during Ben Straub’s (GSK) and Eli Miller’s (Atorus) presentation called “Closing the Gap: Creating an End to End R Package Toolkit for the Clinical Reporting Pipeline.” At the end of the talk, Eli welcomed the community to the pharmaverse. Borrowing ideology from the tidyverse, the pharmaverse is a new group of packages developed by various pharmaceutical companies to support open-source clinical workflows.
The pharmaverse demonstrates the recognition of the best programming language (R) and the best model (open source) for the creation and adoption of industry-standard tools in pharma. Representatives across GSK, Atorus, Roche, Janssen, and Novo Nordisk promote the collaborative development of curated open-source R packages for clinical reporting usage in pharma.
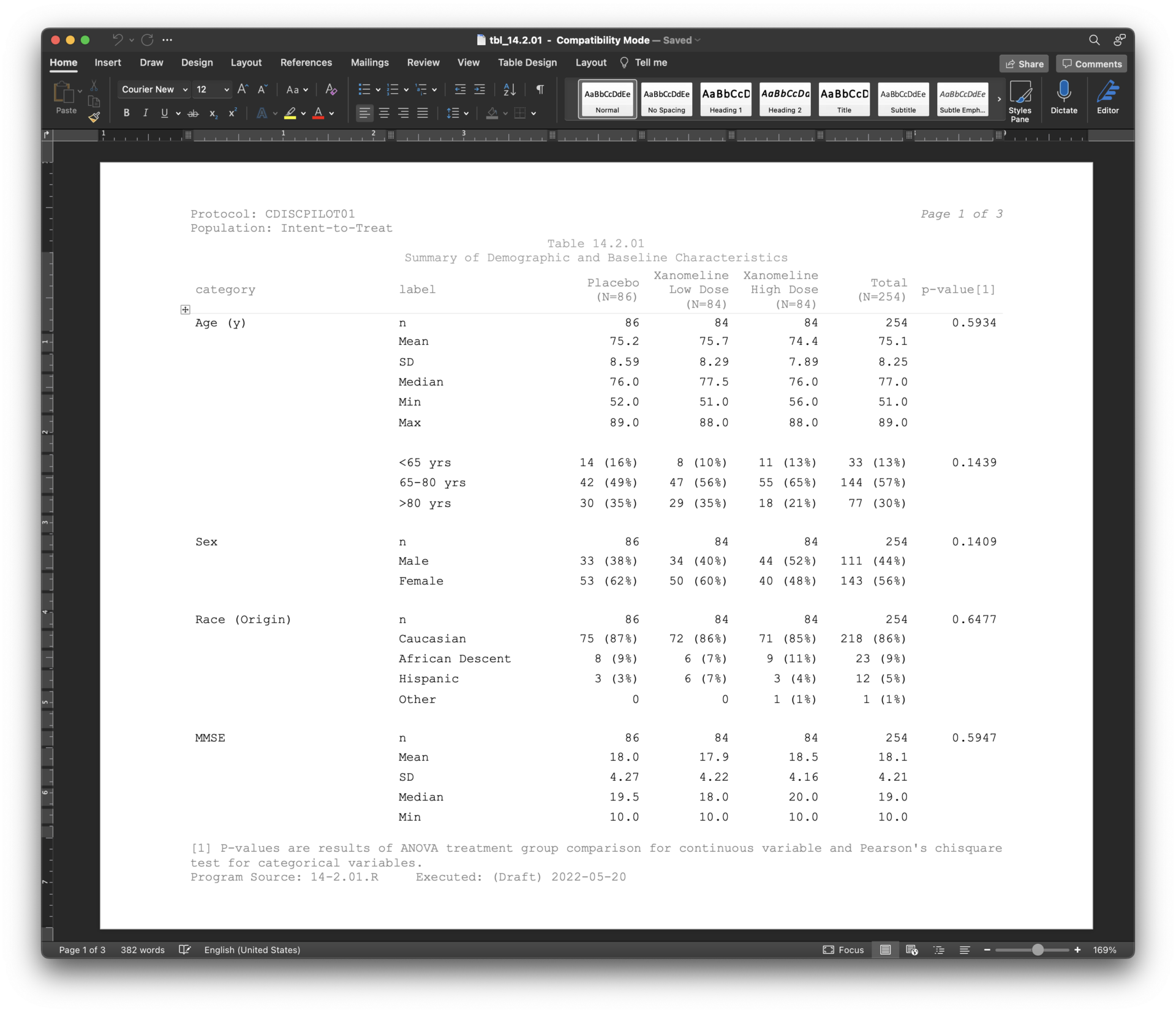
The pharmaverse started as a shared vision of Michael Rimler (GSK), Ross Farrugia (Roche), Mike Stackhouse (Atorus), and Sumesh Kalappurakal (Janssen) to provide the industry with a “pharma stack of open source R packages to enable clinical reporting (from CRF to eSubmission)”. By 2021, several pharmaceutical companies had already started releasing open-source R packages, such as Merck’s r2rtf, Atorus’ pharmaRTF and Tplyr, and Roche’s rtables. The inspiration of pharmaverse was the motivation that much of the work becomes redundant across companies when working on standardized clinical workflows. For instance, one of the most critical standards for clinical trial submission is the Analysis Data Model (ADaM). ADaM standards outline how to create analysis datasets and associated metadata. This allows a statistical programmer to generate figures, listings, and tables while ensuring traceability. Reviewers can assess and approve a submission more quickly. To address the strict standards of the Federal Drug Administration, Roche and GSK joined forces to create the ADaM in R Asset Library package, admiral. Admiral provides a toolbox of reusable functions and utilities with dplyr-like syntax to prep ADaM datasets. It can be used alongside other tidyverse packages so that programmers can build ADaMs according to their varying analysis needs.
Other packages include NEST, a set of R packages for streamlining the generation of interactive analyses, and metacore, for using dataset specification metadata within an R session. Janssen R&D, Merck, and other pharmaceutical companies joined the effort in 2020 and 2021 to contribute code publicly to CRAN for packages they use internally, such as Merck’s pkglite and Roche’s rtables.
In 2021, Roche gave a workshop at R in Pharma called “Clinical Trials Data Analysis at Roche,” highlighting many of the packages contributed to the pharmaverse. In 2022, more packages were added to the pharmaverse, such as tfrmt, a GSK package that provides a language for defining display-related metadata. This package and other pharmaverse packages were featured at the rstudio::conf(2022) workshop, Clinical Reporting in R.
While many of these packages began with a small set of developers, the pharmaverse is open to contributions. Over 100 developers have contributed code to the various packages. A package’s admission into the pharmaverse provides an option for those looking to avoid duplicating solutions. The open-source spirit ensures a fair and representative voice is given across the industry, enabling:
- More robust solutions with shared development and maintenance efforts
- Acceleration of insights due to the pooling of resources
- Unified solutions due to collaboration between organizations
- Increased transparency, as code is released under a permissive license
- Attraction of the next generation of great software developers and data scientists to Pharma
The pharmaverse is an incredible demonstration of the growth and evolution of open source in clinical trials. Its Slack channel, made up of over a thousand members, is available to anyone keen on advancing the development of the pharmaverse.
We at Posit believe the future of pharma is open source, and we also love to contribute to these efforts. Rich Iannone, a developer at Posit, is an active contributor to the R Consortium’s R Tables for Regulatory Submission Working Group and maintains the gt package. With gt, clinical programmers can prepare and customize tables using R. Users write code once and render it to RTF, HTML, LaTeX, and other outputs. The package has been in continuous development at Posit for over three years, and we incorporate features tailored to the pharma industry.
More than just providing technical functionality, open source offers innumerable opportunities for collaboration between the best minds in the field. These efforts to join forces together across the industry are historical. “When you’re thinking of those medicines, when you’re thinking of those chemistries, and how we are innovating in therapeutics, that is where a lot of…desire is to innovate,” says
RStudio Pharma Meetup Series: Data-as-a-Product, A data science framework for data collaboration
Editor’s note: We have edited this post with clarity on the organizations, roles, and descriptions mentioned within the article to provide a more accurate and informative reading experience.
Learn more
We believe this work is just beginning, and we look forward to what’s to come.
- Join us on November 29th as a team from AstraZeneca shares their experience growing an R community at their company.
- Please find out more about how open source changes drug development on our Pharma page.
- Check out a recent ASA paper by Ross Farrugia (Roche) and Sumesh Kalappurakal (Janssen) that gives a wonderful overview of the pharmaverse: Welcome to the pharmaverse!
There were many talks and workshops on the pharmaverse at the 2022 R in Pharma conference. You can see many on YouTube. Highlights include:
Workshops
- Adrian Waddell (Roche) and Gabriel Becker’s (Roche) workshop on the rtable package for clinical trial outputs
- Christina Fillmore (GSK), Ellis Hughes (GSK), and Thomas Neitmann (Roche) on a modern approach to generate ADaMs and TLFs
- Dinakar Kulkarni (Roche) and Ben Straub (GSK) on implementing CI/CD for R packages
- Mike Stackhouse (Atorus) workshop on Drill Down Summary Tables in Shiny with Tplyr
Talks
- Ross Farrugia (Roche) delivering a keynote on the pan-company pharmaverse he co-founded
- Christina Fillmore (GSK) on “Why do I spend all my life formatting tables?!”
- Ning Leng (Roche) and Hye Soo Cho (FDA) co-presenting on a pilot R submission and a career guidance panel
- Daniel Sabanés Bové (Roche) discussing the team he built to catalyze statistical engineering at Roche & on the panel for R governance
- Coline Zeballos (Roche) and Doug Kelkhoff (Roche) talking about how they revolutionized the validation of R packages at Roche
- Will Harris (Genentech) on a well-received STDM checks package
- Dinakar Kulkarni (Roche), Ben Straub (GSK), and Craig Gower-Page (Roche) talking about CI/CD for the pharmaverse
- Kieran Martin (Roche) on an upcoming Coursera course that will help bridge the gap between data science and late-stage drug development
